Biomarker multiplexing
A major focus of the lab is the development and validation of reliable quantitative prognostic and predictive biomarkers.
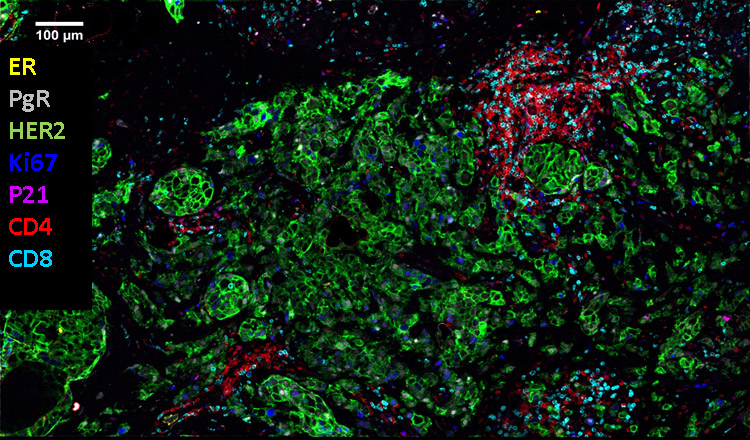 A multiplexed composite image of breast cancer tissue showing co-localization of seven biomarkers.
A multiplexed composite image of breast cancer tissue showing co-localization of seven biomarkers.
Two multiplexing technology platforms are being used and available for all researcher and clinical collaborators: Cell DIVE (Leica Micrsosystems) and Phenocycle Fusion (Akoya Biosciences)
While CellDIVE platform (Figure 5a) uses cyclic immunofluorescence with compatible commercially available antibodies, the Akoya’s platform (Figures 5 b) uses a similar approach but with proprietary barcoding technology and custom antibody panels.
Both systems, while using different approaches, allow for co-localization of up to 100+ biomarkers on a single cell providing exciting possibilities for research on treatment outcome and precision medicine.

View a plain-text version of infographic
Biomarker multiplexing systems
CellDIVE imager (Leica Biosystems):
- Antibody agnostic stain-image-bleach cyclic immunofluorecense system
- Step 1: Antigen retrieval, serum blocking
- Step 2: Label section with CV 3, 3.5, 5, 7 conjugated antibodies
- Step 3: Acquire IF image
- Step 4: Photo-induced chemical bleaching
- Step 5: Acquire background image, repeat
PhenoCycler-Fusion System (Akoya):
- Step 1: Slide preparation and panel assembly
- Slide preparation using standard antigen retrieval and serum blocking protocols
- Step 2: Application of oligonucleotide barcoded antibodies
- Each antibody is tagged with a unique oligonucleotide barcode. Antibodies and reagents are proprietary and provided by Akoya in batches of 10 applications
- Step 3: Iterative cycling and imaging
- The platform works by staining the tissue is a single-step and iteratively cycling and imaging fluorescently labeled antibodies across the tissue through a reveal-image-remove cycle
We are involved in a number of collaborative projects developing and testing biomarker panels for breast, lung, and lymphoma cancers. We have collaborated in the past on ductal carcinoma in situ (DCIS), immunotherapies for ovarian cancer, immune checkpoint inhibitors in breast cancer and immunotherapies in hypermutant cancers including brain and lung cancers.
Highlights of current projects include a collaboration with the OICR Adaptive Oncology program, where we have studied a collection of whole-mount processed breast cancer specimens using biomarker multiplexing and found large levels of intra-tumoural heterogeneity. There we identified various clusters of cancer-associated fibroblasts, and measured if certain groups are more prevalent in breast cancer subtypes (Figure 6).

Using quantitative methods, developed by our team and commercial platforms that measure biomarker signal we are able to define the extent of cancer heterogeneity by characterizing each clone, and investigate potential functional relationships between different biomarkers. This information can be used to delineate further subtypes of cancers for developing treatments that can target heterogeneous tumours.


